NanoString nCounter Max
Breaking news from our Digital Platform for Direct Molecular Detection with Nanostring nCounter® MAX Analysis System:
Our recently acquired equipment nCounter MAX from Nanostring offers a series of advantages over the previous version (SPRINT Profiler), and allows us to improve turn around times by eliminating the main bottlenecks in sample processing and obtaining data and the corresponding results for your project or study. In addition, it allows detecting small structural changes in folding and eliminates the synthesis, amplification, and preparation of cDNA libraries to reduce technical variations in the assay aiming to reduce the need for experimental replicates.
The nCounter® Assay System allows hundreds of mRNAs, miRNAs, SNVs or CNVs to be analyzed directly by direct digital molecular detection, in a single reaction in the absence of enzymes (no reverse transcription or amplification). It is a system of high sensitivity and reproducibility, with a huge multiplexing capacity (up to 800 genes in the same reaction), which not only reduces the number of necessary reactions, but also saves the amount of RNA/DNA that is required for the test.
Technology
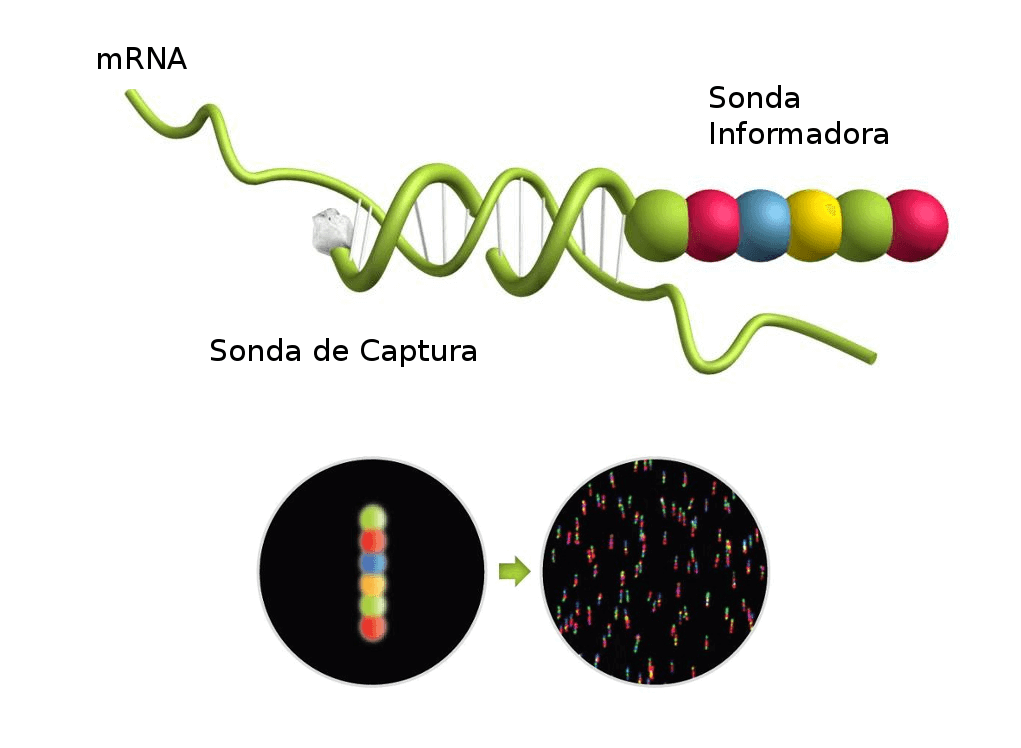
The nCounter Analysis System utilizes a novel digital count through a colour barcode technology for direct multiplexed measurement.
The technology uses molecular "barcodes" and single molecule imaging for the direct hybridization and detection of hundreds of unique transcripts in a single reaction.
Each color-coded barcode is attached to a single target-specific probe corresponding to a molecule of interest. You will obtain the real number of target molecules presents in a given sample.

Technology main advantages
- Time-effective: no enzymatic reaction is required (no amplification / no cDNA production / no library preparation). This minimize technical variation and therefore the generation of biased result.
- Low sample input: it saves on the required sample input amount.
- High precision, sensibility and reproducibility: higher precision than qPCR with no technical replicates required (replicas correlation >0.99). Precision and reproducibility is maintained even at low expression levels. This leads for more precise downstream analysis.
- Versatility: it can work with difficult samples. A wide range of sample types and quality are suitable to be analysed with this technology. It is possible to obtain good results from FFPE samples or directly from cell lysates (without previous RNA extraction).
- Multiplexing: you can analyse hundreds of targets (up to 800) in a single tube. Possibility of using pre-designed panels or building custom panels with the targets of interest.
- Simple data analysis: data output is generated in csv format. Data can be easly exported and analysed with third party software packages.
Applications
Gene expression
Nanostring technology has a great number of potential applications in different areas of the biomedical research field. There are many pre-build panels (12 samples) available as well as the chance of ordering custom panels with your targets of interest (up to 800 targets). Most popular panels are:
- Cancer related pre-built panels:
- Gene expression panels containing genes implicated in oncogenic processes formation.
- Gene expression panels containing genes related with the immune response in different types of cancer.
- Gene expression panels containing genes involved in tumour progression.
- Gene expression fusion genes panels focused on the diagnostic of some types of tumours.
- Pre-built panels related to the expression of genes involved in immune, inflammatory and neurodegenerative pathways in human, mouse and rat.
- PlexSet technology (sample multiplexing): custom panels up to 96 genes and 96 samples per cartridge (12, 24, 48, 72 and 96 samples configuration). Cost-effective technology, perfect for signature validation.
- miRNAs:
- miRNA panel with the 800 more relevant miRNAs in terms of biological function. miRNAs discovery and validation in the same panel.
- miRGE custom panel for the simultaneous analysis of mRNA and miRNA in the same sample.
- lncRNA custom panel for studying up to 800 lncRNAs. It can be combined with mRNA for simultaneous analysis. In addition, it is suitable for investigate lcnRNA-protein interactions (immunoprecipitation).
- Single cell expression panels. They can be used with wide range of sample types: sorting, microdissection or micromanipulation.
- New pre-designed panels for cardiovascular diseases, cell and gene therapy and stem cells.
DNA
- Copy number variation (CNV) analysis panel. High precision at multiallelic CNVs. It works with FFPE samples. Better reproducibility than microarrays platforms (R2=0.99 of Nanostring vs R2=0.91 of microarrays).
- ChIP-String: study of DNA-protein interaction for gene regulation. Excellent option for ChIP-Seq results validation as well as for the study of genes involved in chromatin remodelling and gene expression in a high number of samples.
Proteomics
Protein analysis with Nanostring uses protein of interest specific antibodies labelled through synthetic oligonucleotides with colour barcodes. Nanostring protein detection is well correlated with cytometry assays (R2>0.75).
Nanotring leads simultaneous quantification of proteins and gene expression in the same sample with nCounter® Vantage 3D™ Immune Assays. You can study mRNA and proteins of the cellular surface like receptors, cellular lineage markers or regulatory molecules to investigate changes in cellular type abundances and activation status. In addition, it offers you the oportunity to study mRNA and some intracellular protein levels like cytokines, chemokines and transcription factors involved in initial stages of cellular communication and immune differentiation.
GENvip Nanosting service
Service Includes:
Support
Support with experimental design and protocol development.
Material
Reagents, Codesets and cartridges ordering.
Analysis
Preparation and processing of 12 samples per cartridge, hybridization and imaging.
Results
Results in csv file format containing the raw and normalized data. Support with the data analysis.
Samples
Samples can be extracted from fresh tissues, cell lines or FFPE. Total mRNA, miRNA, DNA or cell lysates can be used depending on the project. Minimum number of samples is 12.
- Sample quality: samples may be free of contaminants (1.7-2.3 A260/280 ratio and 1.8-2.3 A260/230 ratio) and 50% of the fragments should have more than 300 bp.
- Sample amount:
- mRNA: 100 ng of total mRNA (it depends on the panel) with a minimum concentration of 20 ng/µl.
- miRNA: 100-300 ng of total mRNA (minimum concentration of 33 ng/µl).
- CNV y ChIP-String: CNV and ChIP-String: 200-600 ng of DNA (minimum concentration of 29 ng/µl)
GENVIP team is a multidisciplinary group with a cross-sectional nature encompassing different clinic and basic research disciplines. GENVIP team applies last generation molecular and analytical technologies from different disciplines with the aim of improving the clinical management of paediatric patients.
The main goal of GENVIP research team is to deepen the knowledge on pathogenesis of infectious diseases affecting childhood from different perspectives, through a breakthrough ‘omics’ approach. This ‘omics’ approach will allow to identify genetics and transcriptomics biomarkers to define specific diagnostic and prognostic footprints associated to different paediatric infectious disease based on its molecular phenotyping. This will help to acquire a valuable translational knowledge to be used in the clinical practice, contributing to early recognition, selective prevention and management of the pediatric patients. This will also open new lines for personalised treatment of the paediatric patients, contributing to the understanding and prediction of the potentially broad impact of therapeutics and vaccines on the child immune system.